class: center, middle, inverse, title-slide # Implementing proximity based social network analysis using spatsoc ### Alec L. Robitaille, Quinn M.R. Webber and Eric Vander Wal ### 2019-02-07 --- background-image: url("images/ducksandzebras.png") background-position: center background-size: contain # The paradox of being social in spatial context --- # History of spatsoc * ### Linking social behavior and space use * ### Most practical to use animals with GPS collars * ### GPS collars typically used to quantify measures of habitat selection or movement, but not social proximity * ### Webber and Vander Wal review links between social and spatial ecology --- background-image: url("images/evoframe.png") background-position: center background-size: contain # History of spatsoc --- # History of spatsoc: big ideas, big data ## Idea: * #### Individualize social network proximity and habitat selection patterns and determine how these traits influence reproductive success. ## Data: * #### Caribou in Newfoundland, Elk and wolves in Manitoba * #### For example, in Newfoundland, many studies quantifying habitat selection patterns for Newfoundland caribou, but no incorporation of social processes .center[] --- # Informal implementation * ### Need for functions to generate social networks for GPS collared caribou * ### Original “home-made” functions made by Quinn were limited .pull-right[] * #### Inflexible * #### Slow * #### Prone to breaking --- # Formal implementation * #### Early functions simply generated buffers around an individual's temporally discrete GPS fixes and detected whether other individuals were within those buffers .center[] --- # History of spatsoc: putting it to the test .center[  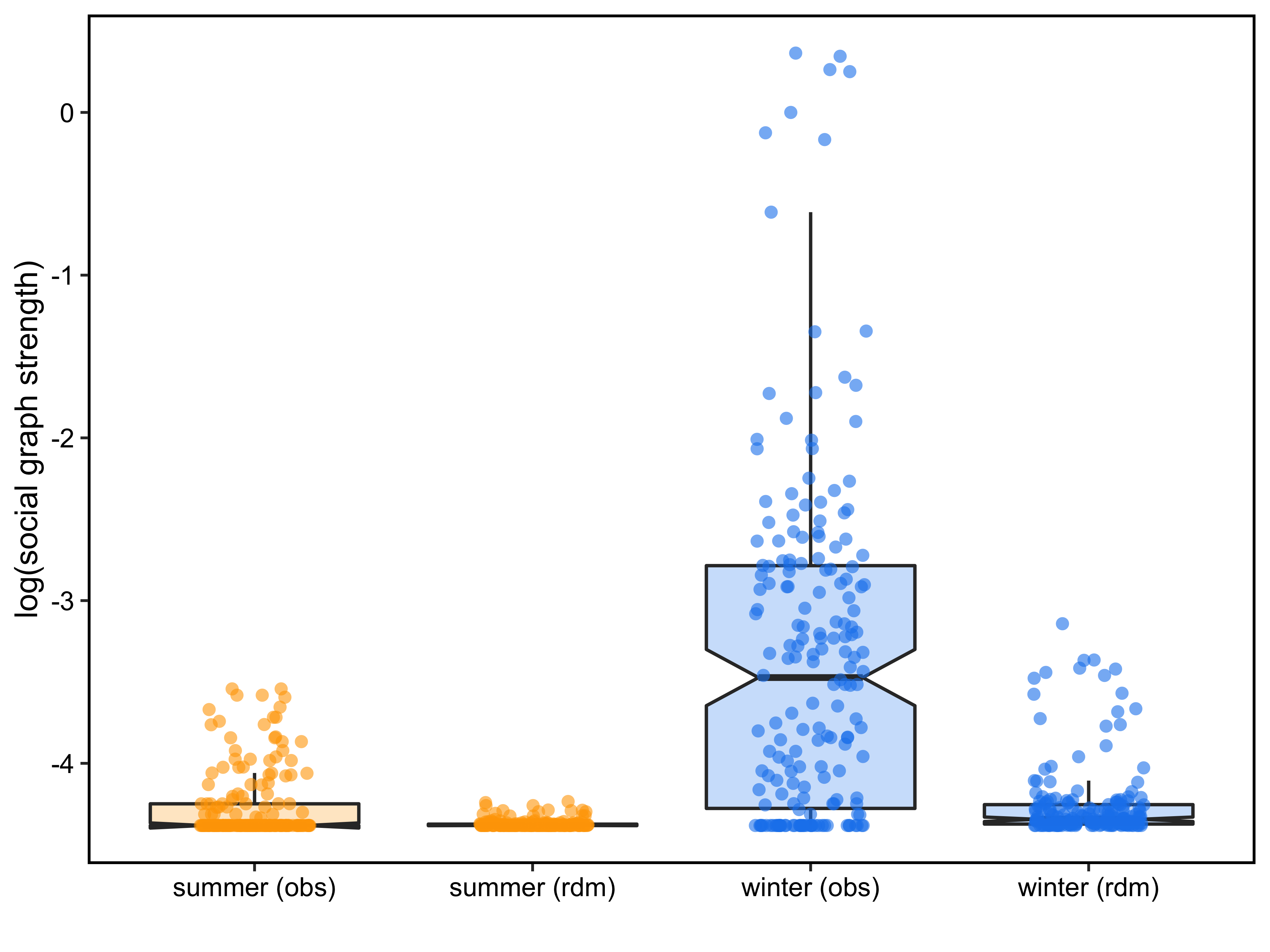 ] ??? spatsoc used to generate proximity based social networks and compared across seasons --- # History of spatsoc: applications * #### Social processes can be relevant for conservation managers: * #### Allee effects * #### Detection-dilution trade-off for predators * #### Detect locations where animals aggregate * #### Determine whether animals more likely to aggregate at certain times of year * #### If survival/reproductive of collared animals is known, can determine whether social association was related to survival .center[] --- # Package basics .pull-left[ <div id="htmlwidget-95ea5ec5b1853010b1a4" style="width:504px;height:504px;" class="grViz html-widget"></div> <script type="application/json" data-for="htmlwidget-95ea5ec5b1853010b1a4">{"x":{"diagram":"digraph {\n\tgraph [rankdir=TB, compound=TRUE, fontname = \"Helvetica\", fontsize = 20, bgcolor = \"#FAFAFA\"]\n node [fontname = \"Helvetica\", fontsize = 22];\n\tscript -> function -> package \n script [label = <Scripts<BR/><FONT POINT-SIZE=\"18\">(useful)<\/FONT>>] \n function [label = <Functions<BR /><FONT POINT-SIZE=\"18\">(reusable)<\/FONT>>] \t\n\tpackage [label = <Packages<BR /><FONT POINT-SIZE=\"18\">(shareable)<\/FONT>>] \n\t}","config":{"engine":"dot","options":null}},"evals":[],"jsHooks":[]}</script> ] .sidebar[ Package checklist: `\(\> \checkmark\)` novel and interesting method `\(\> \Box\)` easy to use, flexible `\(\> \Box\)` extensive documentation `\(\> \Box\)` well tested ] ??? <!-- ## how spatsoc works --> --- # Workflow <div id="htmlwidget-96ebef884d43402a04a9" style="width:800px;height:504px;" class="grViz html-widget"></div> <script type="application/json" data-for="htmlwidget-96ebef884d43402a04a9">{"x":{"diagram":"\n\tdigraph spatsoc_functions {\n\tgraph [rankdir=TB, compound=TRUE, fontname = \"Helvetica\", bgcolor = \"#FAFAFA\"]\n node [fontname = \"Helvetica\"];\n\n\t# functions\n#\tfunction [shape = \"rectangle\"]\n\tspgrp [label = \"spatial grouping\", shape = \"rectangle\"]\n\ttgrp [label = \"temporal grouping\", shape = \"rectangle\"]\n\trand [label = \"randomizations\", shape = \"rectangle\"]\n\n\t# input\n#\tdata [shape = \"diamond\"]\n\tinput [label = \"telemetry data\", shape = \"diamond\"]\n\n\t# output\n#\toutput [shape = \"ellipse\"]\n\tobs [label = \"observed network\", shape = \"ellipse\"]\n\trdm [label = \"randomized networks\", shape = \"ellipse\"]\n\n\n#\tsubgraph cluster_workflow {\n# label= \"Workflow\" labeljust=\"l\";\n\t{rank=same; rdm; obs}\n\n\tinput -> tgrp -> spgrp -> obs\n\tspgrp -> rand -> rdm\n\t#}\n\n#\tsubgraph cluster_legend {\n# label= \"Legend\" labeljust=\"l\";\n#\tdata -> function -> output\n#\t}\n\t}\n\t","config":{"engine":"dot","options":null}},"evals":[],"jsHooks":[]}</script> --- # Temporal grouping ```r group_times(DT, datetime = c('idate', 'itime'), threshold = '5 minutes') ``` .pull-left[ * grouped by "nearest" * threshold must be divisible by 24 hours or 60 minutes |idate | itime| minutes| timegroup| |:----------|--------:|-------:|---------:| |2018-12-12 | 01:00:00| 0| 1| |2018-12-12 | 01:02:00| 0| 1| |2018-12-12 | 01:01:00| 0| 1| |2018-12-12 | 03:00:00| 0| 2| |2018-12-12 | 03:02:00| 0| 2| |2018-12-12 | 03:00:00| 0| 2| |2018-12-12 | 05:00:00| 0| 3| |2018-12-12 | 05:01:00| 0| 3| |2018-12-12 | 05:01:00| 0| 3| ] .fixed-pos-r[ <div id="htmlwidget-1ddad42428ebf81e3a09" style="width:425px;height:504px;" class="grViz html-widget"></div> <script type="application/json" data-for="htmlwidget-1ddad42428ebf81e3a09">{"x":{"diagram":"\n\tdigraph spatsoc_functions {\n\tgraph [rankdir=TB, compound=TRUE, fontname = \"Helvetica\", bgcolor = \"#FAFAFA\"]\n node [fontname = \"Helvetica\"];\n\n\t# functions\n#\tfunction [shape = \"rectangle\"]\n\tspgrp [label = \"spatial grouping\", shape = \"rectangle\"]\n\ttgrp [label = \"temporal grouping\", shape = \"rectangle\", color=\"red\"]\n\trand [label = \"randomizations\", shape = \"rectangle\"]\n\n\t# input\n#\tdata [shape = \"diamond\"]\n\tinput [label = \"telemetry data\", shape = \"diamond\"]\n\n\t# output\n#\toutput [shape = \"ellipse\"]\n\tobs [label = \"observed network\", shape = \"ellipse\"]\n\trdm [label = \"randomized networks\", shape = \"ellipse\"]\n\n\n#\tsubgraph cluster_workflow {\n# label= \"Workflow\" labeljust=\"l\";\n\t{rank=same; rdm; obs}\n\n\tinput -> tgrp -> spgrp -> obs\n\tspgrp -> rand -> rdm\n\t#}\n\n#\tsubgraph cluster_legend {\n# label= \"Legend\" labeljust=\"l\";\n#\tdata -> function -> output\n#\t}\n\t}\n\t","config":{"engine":"dot","options":null}},"evals":[],"jsHooks":[]}</script> ] --- # Spatial grouping .center[[`data.table`](https://github.com/Rdatatable/data.table) + GIS operations + [`igraph`](https://github.com/igraph/rigraph)] .pull-left[ |function | operation | |:----------|--------:| |`group_pts` | distance matrix | |`group_lines` | line intersection | |`group_polys` | polygon overlap | ] .fixed-pos-r[ <div id="htmlwidget-abdeb135a3aac15771b5" style="width:425px;height:504px;" class="grViz html-widget"></div> <script type="application/json" data-for="htmlwidget-abdeb135a3aac15771b5">{"x":{"diagram":"\n\tdigraph spatsoc_functions {\n\tgraph [rankdir=TB, compound=TRUE, fontname = \"Helvetica\", bgcolor = \"#FAFAFA\"]\n node [fontname = \"Helvetica\"];\n\n\t# functions\n\tspgrp [label = \"spatial grouping\", shape = \"rectangle\", color=\"red\"]\n\ttgrp [label = \"temporal grouping\", shape = \"rectangle\"]\n\trand [label = \"randomizations\", shape = \"rectangle\"]\n\n\t# input\n\tinput [label = \"telemetry data\", shape = \"diamond\"]\n\n\t# output\n\tobs [label = \"observed network\", shape = \"ellipse\"]\n\trdm [label = \"randomized networks\", shape = \"ellipse\"]\n\n\n\t{rank=same; rdm; obs}\n\n\tinput -> tgrp -> spgrp -> obs\n\tspgrp -> rand -> rdm\n\t}\n\t","config":{"engine":"dot","options":null}},"evals":[],"jsHooks":[]}</script> ] --- # Spatial grouping: points ```r group_pts(DT, threshold = 5, id = 'ID', coords = c('X', 'Y'), timegroup = 'timegroup') ``` .pull-left[  ] .fixed-pos-r[ <div id="htmlwidget-d60dcf8a46508a53a9ee" style="width:425px;height:504px;" class="grViz html-widget"></div> <script type="application/json" data-for="htmlwidget-d60dcf8a46508a53a9ee">{"x":{"diagram":"\n\tdigraph spatsoc_functions {\n\tgraph [rankdir=TB, compound=TRUE, fontname = \"Helvetica\", bgcolor = \"#FAFAFA\"]\n node [fontname = \"Helvetica\"];\n\n\t# functions\n\tspgrp [label = \"spatial grouping\", shape = \"rectangle\", color=\"red\"]\n\ttgrp [label = \"temporal grouping\", shape = \"rectangle\"]\n\trand [label = \"randomizations\", shape = \"rectangle\"]\n\n\t# input\n\tinput [label = \"telemetry data\", shape = \"diamond\"]\n\n\t# output\n\tobs [label = \"observed network\", shape = \"ellipse\"]\n\trdm [label = \"randomized networks\", shape = \"ellipse\"]\n\n\n\t{rank=same; rdm; obs}\n\n\tinput -> tgrp -> spgrp -> obs\n\tspgrp -> rand -> rdm\n\t}\n\t","config":{"engine":"dot","options":null}},"evals":[],"jsHooks":[]}</script> ] --- # Spatial grouping: lines ```r group_lines(DT, threshold = 50, projection = utm, id = 'ID', coords = c('X', 'Y'), timegroup = 'timegroup', sortBy = 'datetime') ``` .pull-left[  ] .fixed-pos-r[ <div id="htmlwidget-f941f8dd4c9d36e7d619" style="width:425px;height:504px;" class="grViz html-widget"></div> <script type="application/json" data-for="htmlwidget-f941f8dd4c9d36e7d619">{"x":{"diagram":"\n\tdigraph spatsoc_functions {\n\tgraph [rankdir=TB, compound=TRUE, fontname = \"Helvetica\", bgcolor = \"#FAFAFA\"]\n node [fontname = \"Helvetica\"];\n\n\t# functions\n\tspgrp [label = \"spatial grouping\", shape = \"rectangle\", color=\"red\"]\n\ttgrp [label = \"temporal grouping\", shape = \"rectangle\"]\n\trand [label = \"randomizations\", shape = \"rectangle\"]\n\n\t# input\n\tinput [label = \"telemetry data\", shape = \"diamond\"]\n\n\t# output\n\tobs [label = \"observed network\", shape = \"ellipse\"]\n\trdm [label = \"randomized networks\", shape = \"ellipse\"]\n\n\n\t{rank=same; rdm; obs}\n\n\tinput -> tgrp -> spgrp -> obs\n\tspgrp -> rand -> rdm\n\t}\n\t","config":{"engine":"dot","options":null}},"evals":[],"jsHooks":[]}</script> ] --- # Spatial grouping: polygons ```r group_polys(DT, area = FALSE, 'mcp', list(percent = 95), projection = utm, id = 'ID', coords = c('X', 'Y')) ``` .pull-left[ * binary or proportional area overlap  ] .fixed-pos-r[ <div id="htmlwidget-263a2446efc82292e080" style="width:425px;height:504px;" class="grViz html-widget"></div> <script type="application/json" data-for="htmlwidget-263a2446efc82292e080">{"x":{"diagram":"\n\tdigraph spatsoc_functions {\n\tgraph [rankdir=TB, compound=TRUE, fontname = \"Helvetica\", bgcolor = \"#FAFAFA\"]\n node [fontname = \"Helvetica\"];\n\n\t# functions\n\tspgrp [label = \"spatial grouping\", shape = \"rectangle\", color=\"red\"]\n\ttgrp [label = \"temporal grouping\", shape = \"rectangle\"]\n\trand [label = \"randomizations\", shape = \"rectangle\"]\n\n\t# input\n\tinput [label = \"telemetry data\", shape = \"diamond\"]\n\n\t# output\n\tobs [label = \"observed network\", shape = \"ellipse\"]\n\trdm [label = \"randomized networks\", shape = \"ellipse\"]\n\n\n\t{rank=same; rdm; obs}\n\n\tinput -> tgrp -> spgrp -> obs\n\tspgrp -> rand -> rdm\n\t}\n\t","config":{"engine":"dot","options":null}},"evals":[],"jsHooks":[]}</script> ] --- # Spatial grouping all three spatial grouping functions internally use the same logic: 1. use GIS operations to build a logical matrix 1. use `igraph` to calculate the connected components .center[] --- # Flexible thresholds .pull-left[ spatiotemporal grouping with spatsoc is flexible to different user data: 1. Temporal thresholding can be a value on the scale of minutes, hours or days 2. Spatial threshold can be 0 for simple overlap and intersection or any positive value for buffered overlap .warn[ Note that this flexibility comes with a requirement for system specific knowledge and/or sensitivity analysis. ] ] .sidebar[ Package checklist: `\(\> \checkmark\)` novel and interesting method `\(\> \checkmark\)` easy to use, flexible `\(\> \Box\)` extensive documentation `\(\> \Box\)` well tested ] ??? e.g. this warn is a TODO for us --- # Social network analysis ```r randomizations(DT, type = 'trajectory', ...) get_gbi(DT = DT, group = 'group', id = 'ID') ``` .pull-left[  | | A| B| C| D| E| F| |:--|--:|--:|--:|--:|--:|--:| |1 | 1| 1| 1| 0| 1| 1| |2 | 1| 1| 1| 0| 1| 1| |3 | 0| 0| 0| 1| 0| 0| |4 | 0| 0| 0| 1| 0| 0| ] .fixed-pos-r[ <div id="htmlwidget-5d13f5217daa2a76b60c" style="width:425px;height:504px;" class="grViz html-widget"></div> <script type="application/json" data-for="htmlwidget-5d13f5217daa2a76b60c">{"x":{"diagram":"\n\tdigraph spatsoc_functions {\n\tgraph [rankdir=TB, compound=TRUE, fontname = \"Helvetica\", bgcolor = \"#FAFAFA\"]\n node [fontname = \"Helvetica\"];\n\n\t# functions\n\tspgrp [label = \"spatial grouping\", shape = \"rectangle\"]\n\ttgrp [label = \"temporal grouping\", shape = \"rectangle\"]\n\trand [label = \"randomizations\", shape = \"rectangle\", color=\"red\"]\n\n\t# input\n\tinput [label = \"telemetry data\", shape = \"diamond\"]\n\n\t# output\n\tobs [label = \"observed network\", shape = \"ellipse\"]\n\trdm [label = \"randomized networks\", shape = \"ellipse\"]\n\n\n\t{rank=same; rdm; obs}\n\n\tinput -> tgrp -> spgrp\n\tspgrp -> obs [color=\"red\"]\n\tspgrp -> rand\n\trand -> rdm\t[color=\"red\"]\n\t}\n\t","config":{"engine":"dot","options":null}},"evals":[],"jsHooks":[]}</script> ] --- background-image: url("images/ropensci.png") background-position: center background-size: contain # rOpenSci software review <!-- * Editors, reviewers * Standard of rOpenSci: documentation, ease of use * What did the reviewers comment on/ask for improvement --> ??? share this: https://github.com/ropensci/onboarding/issues/237 and this: https://ropensci.org/blog/2018/12/04/spatsoc/ --- # rOpenSci software review .pull-left[ * common goal of improving the package * emphasize extensive testing, continuous integration and user accessibility ] .sidebar[ Package checklist: `\(\> \checkmark\)` original and useful method `\(\> \checkmark\)` easy to use, flexible `\(\> \checkmark\)` extensive documentation `\(\> \checkmark\)` well tested ] .fixed-pos-b[[](https://ropensci.org)] --- # rOpenSci software review .pull-left[] .sidebar[ Package checklist: `\(\> \checkmark\)` original and useful method `\(\> \checkmark\)` easy to use, flexible `\(\> \checkmark\)` extensive documentation `\(\> \checkmark\)` well tested ] ??? expectations <!--basic testing example--> <!-- this example --> --- # Next steps: functions * ### dyadic pairing * ### spatial point clustering .center[[](https://github.com/ropensci/spatsoc/issues)] --- # Next steps: applications * #### spatsoc essentially boils down to points in time and space * #### Can be used to determine encounter rates among predators and prey * #### Could also be used to determine proximity of individuals to fixed locations .center[] --- # Conclusion * ### spatsoc is an R package that groups GPS relocations in time and space * ### Primary intention was to generate proximity based social networks, but other use-cases available * ### spatsoc is in revision for publication, but we’re always looking to grow the package and the functions * ### If you have an idea, let us know or open an issue! --- # Acknowledgements .center[ [](weel.gitlab.io)     [](https://ropensci.org) ] .footnote[ This presentation was built using [Yihui Xie's xaringan package](https://github.com/yihui/xaringan). ]